vue-svg-msa
v3.7.3
Published
Vue component to create a sequence multiple alignment in SVG
Downloads
22
Readme
vue-svg-msa
VueJs component to build a svg multiple sequence alignment.
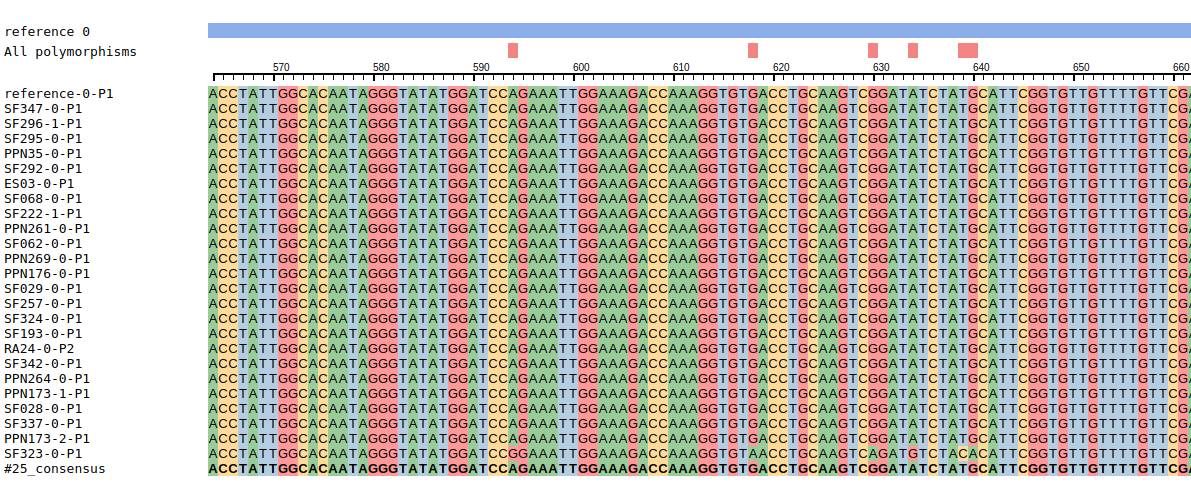
Installation
npm install --save vue-svg-msa npm install --save vue-clipboard2
Usage
in main.js, add:
import VueClipboard from "vue-clipboard2";
Vue.use(VueClipboard);Now you can use it in your component:
import {SvgMsa} from 'vue-svg-msa'
import 'vue-svg-msa/dist/vue-svg-msa.css'<SvgMsa :seqs="seqs">
<SvgMsa
:seqs="seqs"
:type="nt"
:start="start"
:end="end"
:tracks="tracks"
:display-track-tooltip="false"
:metadata="metadata"
:glyphs="glyphs"
:display-glyph-tooltip="true"
:selectedseqs="getselectectids"
:highlight-selection="always"
:offset-x="200"
coloring="auto"
:resolution="sequence"
></SvgMsa>Only the 'seqs' attribute is mandatory. seqs is an array as:
seqs: [
{
seq: 'ATCATCATCATCATCATACTCATTTTTACAT---CATCATACTACATCATCATATACTCATTTTTACATCATC-TCTT',
id: 'seqid1',
name: 'sequence1',
color: 'red',
titleColor: 'blue',
isConsensus: false,
isNode: false
},
...
],Only the 'seq' and 'id' attributes are mandatory. If the 'titleColor' attribute is defined, the name of the sequence is written in this color. The 'color' attribute is the nucleotide background color only applied when the 'coloring' attribute of svg-msa is equal to 'seqcolor'. The sequences for which the attribute isConsensus is true are displayed in bold. The name of the sequences for which the attribute isNode is true can be clicked, and an event named 'select-node' emits the id of the sequence.
type is the composition of the sequences: 'nt' for nucleotides (value by default) or 'aa' for amino acids.
start and end are respectively the start and end positions (1-based) of the sequence regions to be displayed.
To display additional tracks in the top of the svg, complete the tracks array. tracks is an array as:
tracks: [
{
"features": [
{
"positions": [[1,20],[52,80]],
"type": "exon",
"color": "green",
"fill-opacity": "0.5"
},
{
"positions": [[21,51]],
"type": "intron",
"color": "yellow"
}
],
"trackLabel": "exon/intron"
}]If display-track-tooltip is true, display the type and the positions of the feature when hovering with the mouse
coloring allows to colored the nucleotides. The accepted values are 'no', 'seqcolor', 'auto', 'metadata'. It is allowed to mix the values, eg 'seqcolor|metadata' If the value is 'metadata', the styles described in the metadata array are apply to the svg image.
metadata: [
{
label: "Conservation level",
categories: [
{
label: "high level",
style: {
fill: "red",
"fill-opacity": 0.5,
stroke: "black",
},
regions: [
{
id: "seqid1",
ranges: [
[1, 31],
[35, 78],
],
},
{
id: "seqid2",
ranges: [[1, 31]],
},
],
},
],
}, {...}
]In a region (=an element of the 'regions' array), if the 'id' attribute is empty or missing, the regions of all the sequences would be colored. In a region, if the 'ranges' attribute is empty or missing, all the sequence would be colored.
Note: 'label' attributes are not used.
selectedseqs is an array of sequence ids which will be highlighted in the msa.
glyphs is an array used to display glyphs to makes possible to add information to describe the sequences. for now, for all the sequences of a categorie, a colored circle is drawn to the left of the sequence
glyphs: [
{
label: 'resistant/not_resistant',
categories: [
{ label: 'resistant', style: { fill: 'red' }, ids: ['seqid1'] },
{ label: 'not resistant', style: { fill: 'pink' }, ids: ['seqid2', 'seqid3'] }
]
}, ...
]If display-glyph-tooltip is true, display the label of the glyph when hovering the mouse over a circle
resolution allows the values 'sequence' (by default) and 'nt'. If "nt", the rendering quality would be better and hovering the mouse over a nucleotide gives access to its position in the sequence.
highlight-selection: by default, if there are selected sequences, metadata on non selected sequences will appear with less opacity. If the highlight-selection property is set to 'always', metadata on non selected sequences will appear with less opacity even if there is no selected sequences. On the contrary, if the highlight-selection property is set to 'never', metadata will appear always with the same opacity, whatever if the sequence is selected or not.
##Contributors
LIPM Bioinfo Team
- Erika Sallet
- Ludovic Cottret
- Sebastien Carrere
- Ludovic Legrand
- Jerome Gouzy
##Contact
