@rnacentral/fornac
v1.1.10
Published
An RNA secondary structure display component
Downloads
23
Readme
FornaContainer
In many situations, the user interaction is superfluous and the desired goal is
to simply display a secondary structure on a web page. This is a common
scenario in, for example, servers that predict a secondary structure. The
output, a dot-bracket string can simply be added to a FornaContainer object
to display.
Trivial Example
Below is an example of a simple web page which uses a FornaContainer to show
a simple RNA molecule:

The code for creating this web page is rather straightforward. After importing
some necessary javascript files, we create a container using new
FornaContainer("#rna_ss", {'applyForce': false}), passing in #rna_ss as the
id of the div which will hold the container and then populate it with a
structure and sequence using container.addRNA:
<!DOCTYPE html>
<meta charset="utf-8">
This is an RNA container.
<div id='rna_ss'> </div>
This after the RNA container.
<link rel='stylesheet' type='text/css' href='styles/fornac.css' />
<script type='text/javascript' src='scripts/fornac.js'></script>
<script type='text/javascript'>
var container = new FornaContainer("#rna_ss", {'applyForce': false});
var options = {'structure': '((..((....)).(((....))).))',
'sequence': 'CGCUUCAUAUAAUCCUAAUGACCUAU'
};
container.addRNA(options.structure, options);
</script>Cofolded sequences
Display two cofolded sequences using the format of RNAcofold:
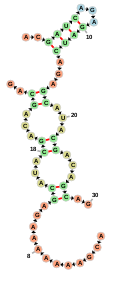
var container = new fornac.FornaContainer("#cofold_ss",
{'applyForce': false, 'allowPanningAndZooming': true, 'initialSize':[500,300]});
var options = {'structure': '..((((...))))...((...((...((..&............))...))...))..',
'sequence': 'ACGAUCAGAGAUCAGAGCAUACGACAGCAG&ACGAAAAAAAGAGCAUACGACAGCAG'
};
container.addRNA(options.structure, options);
container.setSize(); Options
The FornaContainer supports a number of options to allow users to customize how the RNA will be presented.
applyForce
Indicate whether the force-directed layout will be applied to the displayed molecule. Enabling this option allows users to change the layout of the molecule by selecting and dragging the individual nucleotide nodes
allowPanningAndZooming [default=true]
Allow users to zoom in and pan the display. If this is enabled then pressing the 'c' key on the keyboard will center the view.
circularizeExternal [default=true]
This only makes sense in connection with the applyForce argument. If it's
true, the external loops will be arranged in a nice circle. If false, they will
be allowed to flop around as the force layout dictates:
labelInterval [default=10]
Change how often nucleotide numbers are labelled with their number.
Implementation
Each RNA molecule is represented as a JSON file which encodes all of the
information necessary to display it. The example shows a trivial and slightly
modified example. nodeType can be either nucleotide or label or middle,
the latter of which is used only as a placeholder for maintaining an aesthetically
pleasing layout.
The links can be any of basepair (representing a basepair between two
nucleotides), backbone (backbone bond between adjacent nodes), pseudoknot
(pseudoknot, extracted from the specified structure using maximum matching
algorithm), extra (extra links specified by the user), label_link (links
between nucleotides and their nucleotide number labels), fake (invisible
links for maintaining the layout), and fake_fake (invisible links for
maintaining the layout).
This structure is initially created in rnagraph.js starting from a sequence
and dotbracket string.
{
"nodes":
[ {
"name": "A",
"num": 1,
"radius": 5,
"rna": null,
"nodeType": "nucleotide",
"structName": "empty",
"elemType": "e",
"uid": "44edb966-aca9-4058-a6bc-784a34959329",
"linked": false,
"prevNode": null,
"nextNode": null,
"x": 100,
"px": 100,
"y": 100,
"py": 100
},
...
],
"links":
[ {
"source": null,
"target": null,
"linkType": "basepair",
"value": 1,
"uid": "6664a569-5af1-4d86-8ada-d1c00da72a899f87a224-52a0-4ede-a29c-04fddc09e4c4"
},
...
]
}Development
First:
npm install
bower installTo debug:
gulp serveTo build:
gulp buildThe output will be placed in the dist directory. To use fornac in a web page, simply include dist/scripts/fornac.js in your web page.
Acknowledgements
Thanks to Benedikt Rauscher for the Javascript version of the NAView layout and to the creators of VARNA for the java implementation that it's based on.
